Below are selected highlights from the lab's work. A full list of publications is available below.
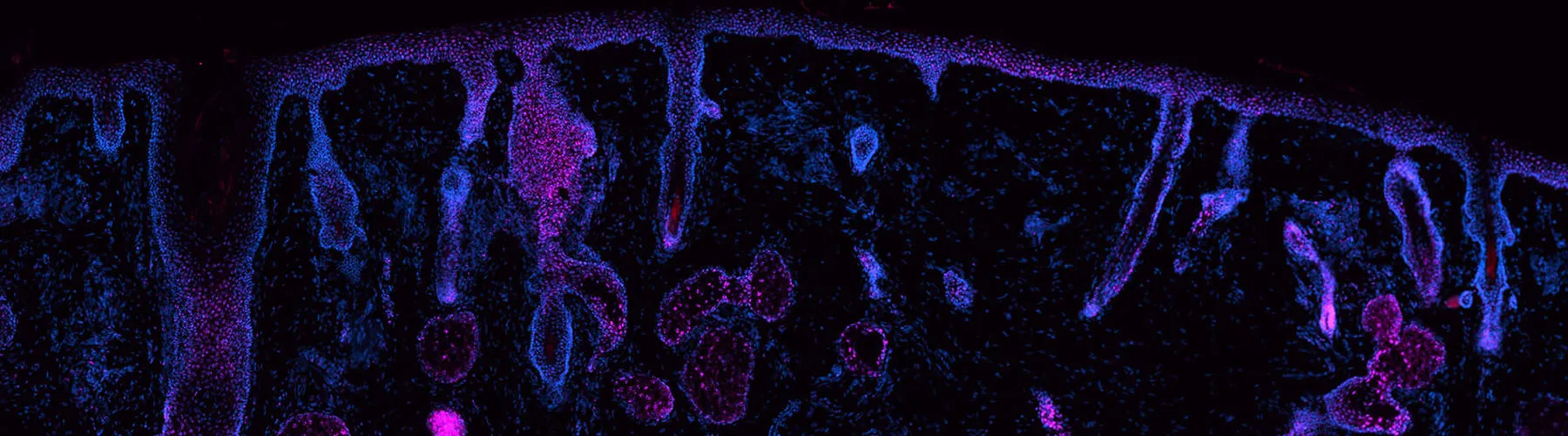
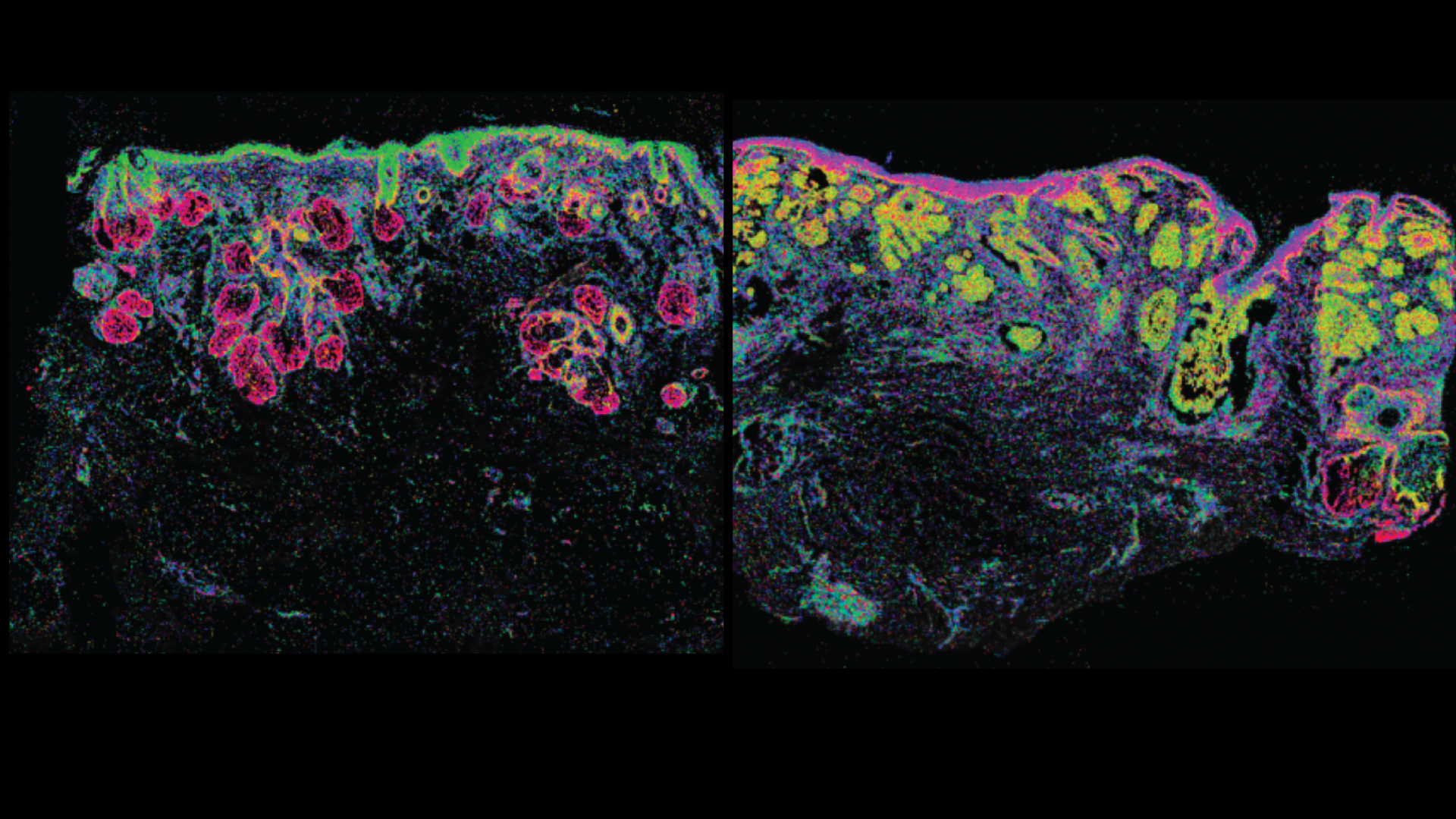
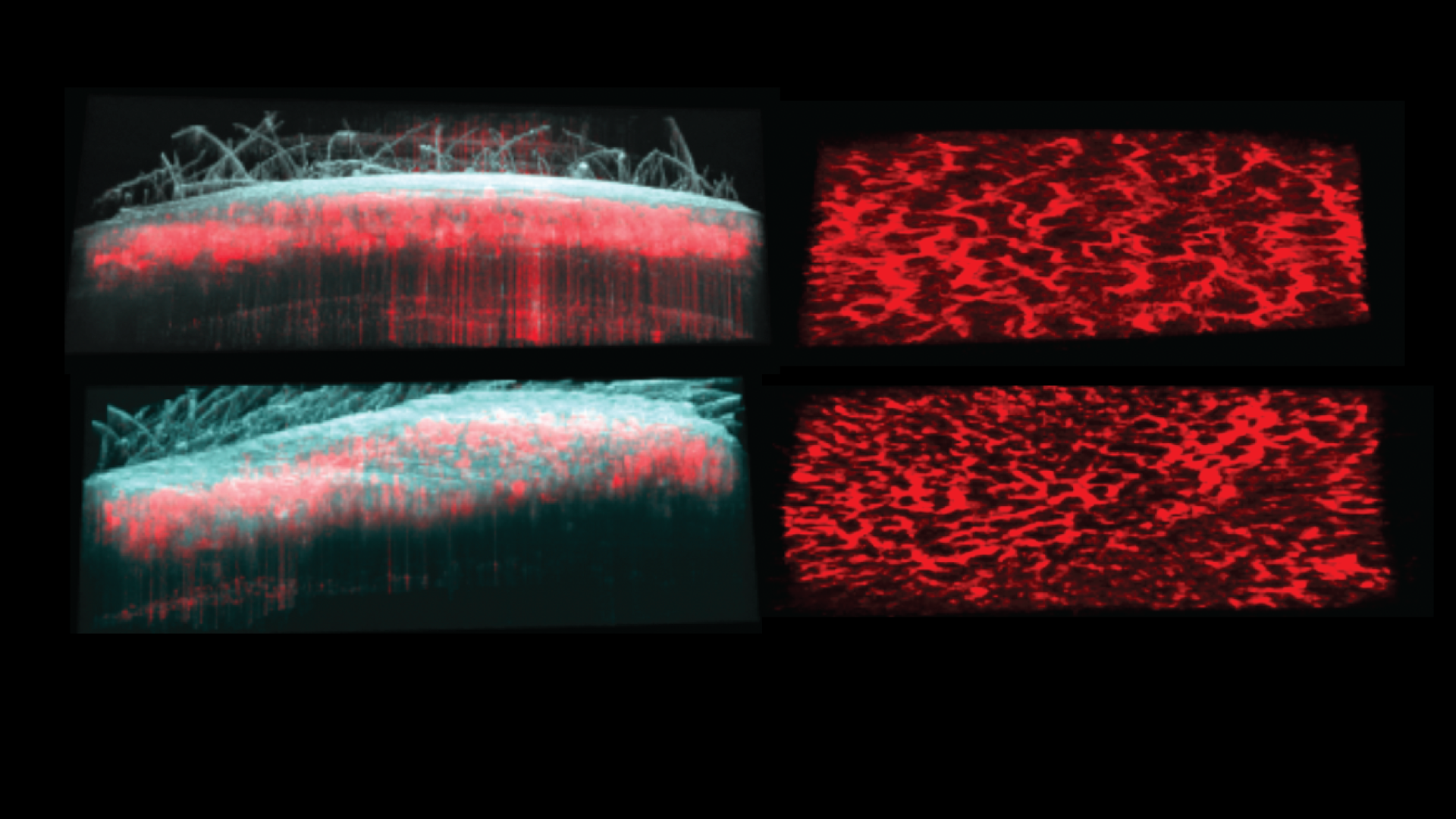
Ganier C, Mazin P, Herrera-Oropeza G, Du-Harpur X, Blakeley M, Gabriel J, Predeus AV, Cakir B, Prete M, Harun N, Darrigrand JF, Haiser A, Wyles S, Shaw T, Teichmann SA, Haniffa M, Watt FM, Lynch MD.
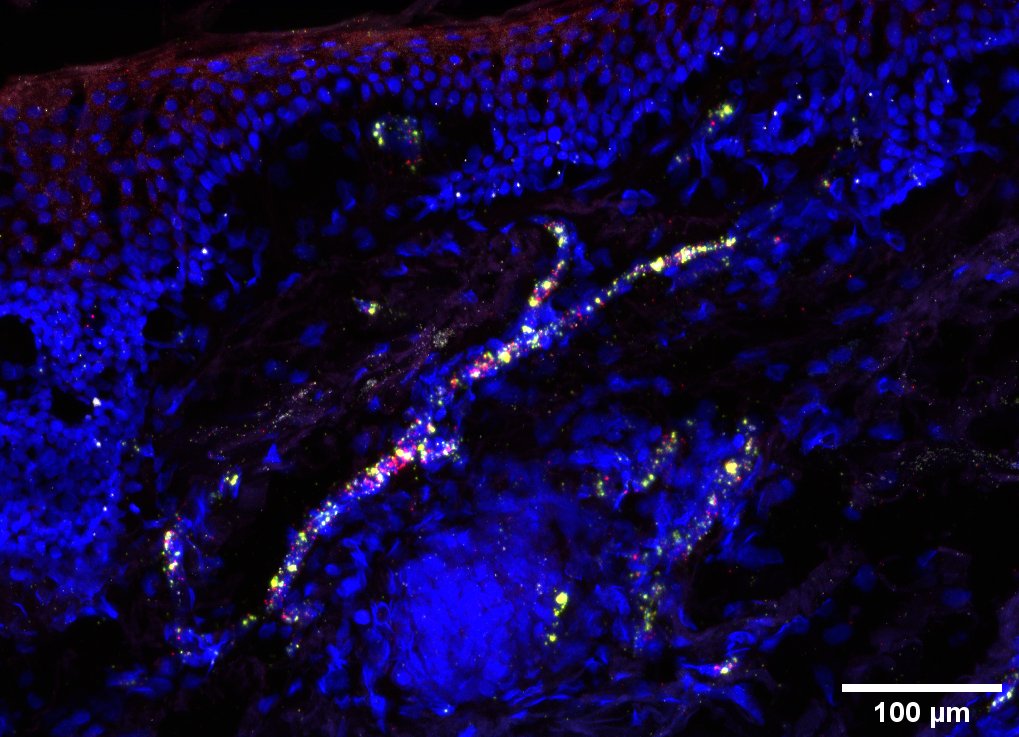
Multiscale spatial mapping of cell populations across anatomical sites in healthy human skin and basal cell carcinoma.
PNAS, 2024.
Pubmed ID: 38165934
This senior-author paper from the Lynch Lab provides unprecedented insight into the cellular architecture of healthy skin and the basal cell carcinoma (BCC) microenvironment. The work creates a multiscale spatial atlas that directly informs the lab's novel cancer prevention strategies.
Reynolds G, Vegh P, Fletcher J,... Lynch MD,...Teichmann S, Watt F, Haniffa M.
Developmental cell programs are co-opted in inflammatory skin disease.
Science, 2021.
Pubmed ID: 33479125
As part of the Human Cell Atlas collaboration, this paper describes the diverse cell types present in healthy skin, eczema, and psoriasis. This foundational atlas serves as a springboard for the Lynch Lab's current work examining different anatomical sites, apatial transcriptomics, and skin cancer.
Du-Harpur X, Arthurs C, Ganier C, Woolf RT, Laftah Z, Lakhan, MK, Salam A, Wan B, Watt FM, Luscombe NM, Lynch MD.
Clinically-relevant vulnerabilities of deep machine learning systems for skin cancer diagnosis.
Journal of Investigative Dermatology, 2020. Pubmed ID: 32931808
Led by the Lynch Lab, this project was the first to show that simple variations in image color balance and rotation can dramatically affect the accuracy of deep learning models for skin cancer diagnosis, highlighting clinically-relevant vulnerabilities in AI-based diagnostic systems.
Philippeos C, Telerman S, Oulès B, Pisco A, Shaw T, Elgueta R, Lombardi G, Driskell R, Soldin M, Lynch MD and Watt FM (joint senior author).
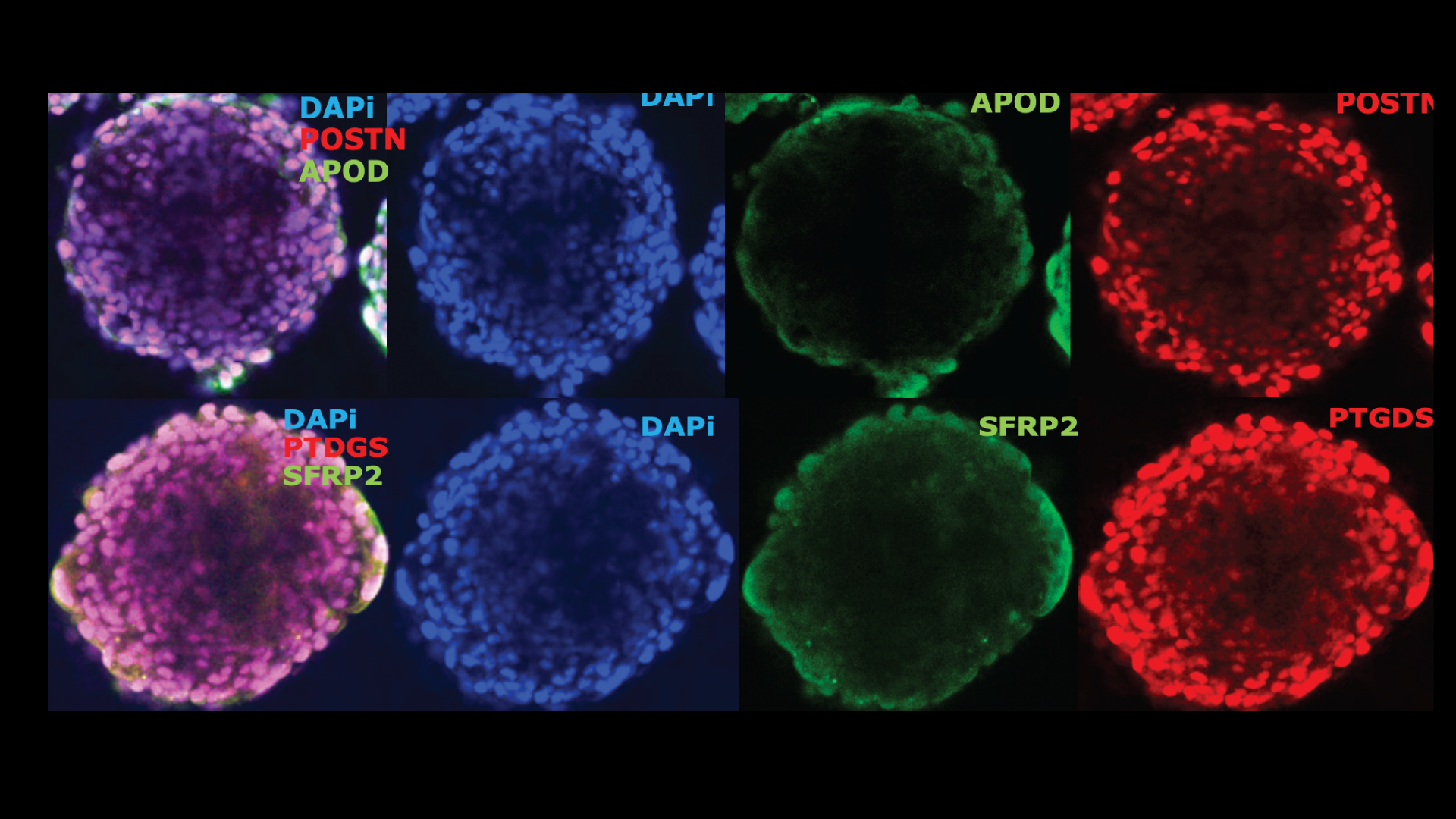
Spatial and single-cell transcriptional profiling identifies functionally distinct human dermal fibroblast subpopulations.
Journal of Investigative Dermatology, 2018. Pubmed ID: 29391249
In this joint senior-author paper, the lab contributed key single-cell sequencing and apatial transcriptomic experiments. The study successfully identified multiple, functionally distinct fibroblast subpopulations in human skin, creating a new and detailed census of fibroblast heterogeneity.
Lynch MD, Lynch CNS, Craythorne E, Liakath-Ali K, Mallipeddi R, Barker JN and Watt FM.
Spatial constraints govern competition of mutant clones in human epidermis.
Nature Communications, 2017. Pubmed ID: 29066762
In this foundational first-author paper, Dr. Lynch demonstrated that sun-exposed, non-cancerous skin contains a high burden of expanded mutant clones. The work revealed that apatial constraints are a major factor governing the competition and selection of these clones within the human epidermis.